For November, it’s that smell of sulfur
We mark the 150th anniversary of Dimitri Mendeleev’s periodic table of chemical elements this year by highlighting elements with fundamental roles in biochemistry and molecular biology. So far, we’ve covered hydrogen, iron, sodium, potassium, chlorine, copper, calcium, phosphorus, carbon, nitrogen, oxygen, manganese and magnesium.
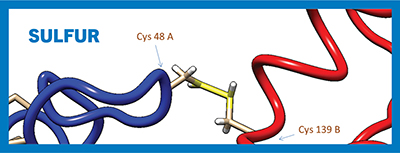 Depiction of the intermolecular disulfide bridge between cysteine residues 48 A and 139 B on opposite chains of the human phenylethanolamine N-methyltransferase dimer.Mikejl921/Wikimedia Common
Depiction of the intermolecular disulfide bridge between cysteine residues 48 A and 139 B on opposite chains of the human phenylethanolamine N-methyltransferase dimer.Mikejl921/Wikimedia Common
In November, families all over the U.S. celebrate Thanksgiving by eating roasted turkey. During digestion, the proteins in that turkey (and other foods) break down into smaller amino acids that are used by the body’s cells to synthesize its own proteins.
Methionine and cysteine — two of the 20 amino acids normally present in the proteins we consume — are our major dietary source of the element sulfur. The organic compound methysulfonylmethane, or MSM, present in vegetables, legumes, herbs and animal products, is another important natural precursor of sulfur.
With chemical symbol S and atomic number 16, sulfur is a reactive nonmetallic element with oxidation states ranging from -2 to +6. Depending on the environment, sulfur compounds may accept or donate electrons. Sulfur reacts with almost all other elements — except for noble gases — and commonly forms polycationic compounds.
Sulfur is created inside massive stars as the product of the nuclear fusion between silicon and helium. By mass, sulfur is the 10th most common element in the universe — present even in many types of meteorites — and the fifth most common element in the Earth. Natural events such as volcanic eruptions, hot spring vents and tidal flats emit abundant sulfur that later accumulates either as pure elemental crystals or as sulfate and sulfide minerals and rocks.
In most ecosystems, the weathering of ore minerals releases stored sulfur that reacts with the oxygen in the air to produce sulfate particles. Sulfate is assimilated by plants and microorganisms and converted into essential biomolecules such as vitamins and proteins, subsequently moving up the food chain. Sulfur is released back onto the land either by decomposition of organic matter or in rainfall. Terrestrial sulfur drains into water systems, where it cycles through marine communities or deposits in deep ocean sediments.
Some anaerobic bacteria in human gut flora can oxidize organic compounds or molecular hydrogen as an energy source and use sulfate instead of oxygen as the final electron acceptor during respiration. These prokaryotes often produce hydrogen sulfide, which causes intestinal gases and flatulence in humans.
In stark contrast, some microorganisms, such as green and purple sulfur bacteria, can use reduced sulfur compounds, and even elemental sulfur, as an energy source to produce sugars by chemosynthesis, using oxygen as the final electron acceptor. Giant tube worms that live on the Pacific Ocean floor rely on these bacteria to feed on seawater sulfide.
Plants and bacteria use environmental sulfate for the synthesis of the nonessential amino acid cysteine. Mammals also can synthesize cysteine from the glycolytic intermediate 3-phosphoglycerate, but they use methionine — an essential amino acid that must be ingested — as the supplier of the sulfur atom. Neighboring cysteine residues in peptide chains can form covalent that contribute to protein flexibility and control protein structure and assembly.
A year of (bio)chemical elements
Read the whole series:
For January, it’s atomic No. 1
For February, it’s iron — atomic No. 26
For March, it’s a renal three-fer: sodium, potassium and chlorine
For April, it’s copper — atomic No. 29
For May, it’s in your bones: calcium and phosphorus
For June and July, it’s atomic Nos. 6 and 7
Breathe deep — for August, it’s oxygen
Manganese seldom travels alone
For October, magnesium helps the leaves stay green
Enjoy reading ASBMB Today?
Become a member to receive the print edition four times a year and the digital edition monthly.
Learn moreGet the latest from ASBMB Today
Enter your email address, and we’ll send you a weekly email with recent articles, interviews and more.
Latest in Science
Science highlights or most popular articles

How scientists identified a new neuromuscular disease
NIH researchers discover Morimoto–Ryu–Malicdan syndrome, after finding shared symptoms and RFC4 gene variants in nine patients, offering hope for faster diagnosis and future treatments.

Unraveling cancer’s spaghetti proteins
MOSAIC scholar Katie Dunleavy investigates how Aurora kinase A shields oncogene c-MYC from degradation, using cutting-edge techniques to uncover new strategies targeting “undruggable” molecules.

How HCMV hijacks host cells — and beyond
Ileana Cristea, an ASBMB Breakthroughs webinar speaker, presented her research on how viruses reprogram cell structure and metabolism to enhance infection and how these mechanisms might link viral infections to cancer and other diseases.

Understanding the lipid link to gene expression in the nucleus
Ray Blind, an ASBMB Breakthroughs speaker, presented his research on how lipids and sugars in the cell nucleus are involved in signaling and gene expression and how these pathways could be targeted to identify therapeutics for diseases like cancer.

Receptor antagonist reduces age-related bone loss in mice
Receptor antagonist reduces bone loss and promotes osteoblast activity in aging mice, highlighting its potential to treat osteoporosis. Read more about this recent JBC paper.

Engineered fusion protein targets kiwifruit pathogen
Synthetic protein selectively kills kiwifruit pathogen, offering a promising biocontrol strategy for agriculture. Read more about this recent JBC paper.

